Home > Press > Chemists Reach from the Molecular to the Real World with Creation of 3-D DNA Crystals
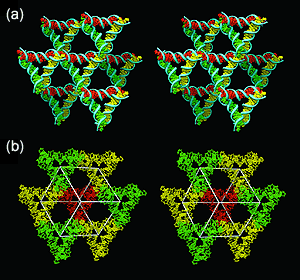 |
| Researchers created 3D DNA structures by using single-stranded sticky ends that link double helices in DNA triangles that point in different directions. |
Abstract:
New York University chemists have created three-dimensional DNA structures, a breakthrough bridging the molecular world to the world where we live. The work, reported in the latest issue of the journal Nature, also has a range of potential industrial and pharmaceutical applications, such as the creation of nanoelectronic components and the organization of drug receptor targets to enable illumination of their 3D structures.
Chemists Reach from the Molecular to the Real World with Creation of 3-D DNA Crystals
Upton, NY | Posted on September 3rd, 2009While scientists, including those involved in this study, have previously designed and built crystal structures, these compositions have been two-dimensional—that is, their axes are on a single plane— and are not the most complete representation of crystals.
To address this limitation, the research team, headed by NYU Chemistry Professor Nadrian Seeman, sought to design and build three-dimensional DNA crystals—a process that requires significant spatial control of the 3D structure of matter. The project also included researchers from Purdue University's Department of Chemistry and the Argonne National Laboratory in Illinois.
To do this, the researchers created DNA crystals by making synthetic sequences of DNA that have the ability to self-assemble into a series of 3D triangle-like motifs. The creation of the crystals was dependent on putting "sticky ends"—small cohesive sequences on each end of the motif—that attach to other molecules and place them in a set order and orientation. The make-up of these sticky ends allows the motifs to attach to each other in a programmed fashion.
Seeman and his colleagues had previously created crystals using this process. However, because these crystals self-assembled on the same plane, they were two-dimensional in composition. In the work reported in Nature, the researchers expanded on the earlier efforts by taking advantage of DNA's double-helix structure to create 3D crystals. The 2D crystals are very small—about 1/1000th of a millimeter—but the 3D crystals are between 1/4 and 1 millimeter, visible to the naked eye.
DNA's double helices form when single strands of DNA—each containing four molecular components called bases, attached to backbone—self-assemble by matching up their bases. The researchers added sticky ends to these double helices, forming single-stranded overhangs to each double helix. Where these overhanging sticky ends were complementary, they bind together to link two double helices. This is a common technique used by genetic engineers, who apply it on a much larger scale. By linking together multiple helices through single-stranded sticky ends, the researchers were able to form a lattice-like structure that extends in six different directions, thereby yielding a 3D crystal.
"With this technique we can organize more matter and work with it in many more ways than we could with 2D crystals," Seeman observed.
A promising avenue for the application of this approach is in nanoelectronics, using components no bigger than single molecules. Currently, such products are built with 2D components. Given the enhanced flexibility that 3D components would yield, manufacturers could build parts that are smaller and closer together as well as more sophisticated in design.
The scientists also expect that they can organize biological macromolecules by attaching them to these crystals. This can help in the development of drugs because macromolecules arranged in crystals can be visualized by a technique known as x-ray crystallography. By adding drugs to these crystals, their interactions with these biological components can be visualized.
X-ray diffraction data were collected from DNA crystals and their iodinated derivatives on beamlines X6A and X25 at the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory in Upton, New York and on beamline ID19 at the Structural Biology Center at Advanced Photon Source in Argonne, Illinois.
Bob Sweet, a biophysicist and the leader of the group that runs NSLS beamline X25, observed, "This is one of the neatest structures I've seen in years. It really connects biotechnology to nanotechnology. We've been helping these folks for over a dozen years, and they really hit the ball out of the park. It's beautiful!"
The research was supported by grants from the National Institute of General Medical Sciences, the National Institutes of Health, the National Science Foundation, the Army Research Office, the Office of Naval Research, and the W.M. Keck Foundation.
####
About Brookhaven National Laboratory
One of ten national laboratories overseen and primarily funded by the Office of Science of the U.S. Department of Energy (DOE), Brookhaven National Laboratory conducts research in the physical, biomedical, and environmental sciences, as well as in energy technologies and national security. Brookhaven Lab also builds and operates major scientific facilities available to university, industry and government researchers. Brookhaven is operated and managed for DOE's Office of Science by Brookhaven Science Associates, a limited-liability company founded by Stony Brook University, the largest academic user of Laboratory facilities, and Battelle, a nonprofit, applied science and technology organization.
For more information, please click here
Contacts:
Kendra Snyder
(631) 344-8191
Mona S. Rowe
(631) 344-5056
Copyright © Brookhaven National Laboratory
If you have a comment, please Contact us.Issuers of news releases, not 7th Wave, Inc. or Nanotechnology Now, are solely responsible for the accuracy of the content.
| Related News Press |
Govt.-Legislation/Regulation/Funding/Policy
![]() Metasurfaces smooth light to boost magnetic sensing precision January 30th, 2026
Metasurfaces smooth light to boost magnetic sensing precision January 30th, 2026
![]() New imaging approach transforms study of bacterial biofilms August 8th, 2025
New imaging approach transforms study of bacterial biofilms August 8th, 2025
![]() Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Possible Futures
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Self Assembly
![]() Diamond glitter: A play of colors with artificial DNA crystals May 17th, 2024
Diamond glitter: A play of colors with artificial DNA crystals May 17th, 2024
![]() Liquid crystal templated chiral nanomaterials October 14th, 2022
Liquid crystal templated chiral nanomaterials October 14th, 2022
![]() Nanoclusters self-organize into centimeter-scale hierarchical assemblies April 22nd, 2022
Nanoclusters self-organize into centimeter-scale hierarchical assemblies April 22nd, 2022
![]() Atom by atom: building precise smaller nanoparticles with templates March 4th, 2022
Atom by atom: building precise smaller nanoparticles with templates March 4th, 2022
Nanomedicine
![]() New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
![]() New imaging approach transforms study of bacterial biofilms August 8th, 2025
New imaging approach transforms study of bacterial biofilms August 8th, 2025
![]() Cambridge chemists discover simple way to build bigger molecules – one carbon at a time June 6th, 2025
Cambridge chemists discover simple way to build bigger molecules – one carbon at a time June 6th, 2025
![]() Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Nanoelectronics
![]() Lab to industry: InSe wafer-scale breakthrough for future electronics August 8th, 2025
Lab to industry: InSe wafer-scale breakthrough for future electronics August 8th, 2025
![]() Interdisciplinary: Rice team tackles the future of semiconductors Multiferroics could be the key to ultralow-energy computing October 6th, 2023
Interdisciplinary: Rice team tackles the future of semiconductors Multiferroics could be the key to ultralow-energy computing October 6th, 2023
![]() Key element for a scalable quantum computer: Physicists from Forschungszentrum Jülich and RWTH Aachen University demonstrate electron transport on a quantum chip September 23rd, 2022
Key element for a scalable quantum computer: Physicists from Forschungszentrum Jülich and RWTH Aachen University demonstrate electron transport on a quantum chip September 23rd, 2022
![]() Reduced power consumption in semiconductor devices September 23rd, 2022
Reduced power consumption in semiconductor devices September 23rd, 2022
Announcements
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Nanobiotechnology
![]() New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
![]() New imaging approach transforms study of bacterial biofilms August 8th, 2025
New imaging approach transforms study of bacterial biofilms August 8th, 2025
![]() Ben-Gurion University of the Negev researchers several steps closer to harnessing patient's own T-cells to fight off cancer June 6th, 2025
Ben-Gurion University of the Negev researchers several steps closer to harnessing patient's own T-cells to fight off cancer June 6th, 2025
![]() Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
|
|
||
|
|
||
| The latest news from around the world, FREE | ||
|
|
||
|
|
||
| Premium Products | ||
|
|
||
|
Only the news you want to read!
Learn More |
||
|
|
||
|
Full-service, expert consulting
Learn More |
||
|
|
||