Home > Press > A better way to read the genome: If your genome was a library, some genes would be 'Choose Your Own Adventure' novels.
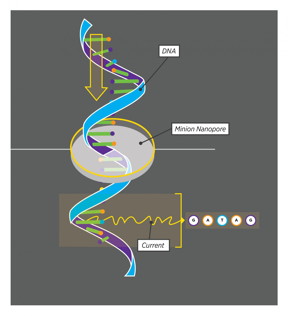 |
| As a strand of DNA moves through the MinION nanopore, the MinION 'sees' five nucleotides at a time. Every set of five nucleotides produces a characteristic current, allowing the MinION to read out the DNA code in a single, continuous stream. CREDIT: Yesenia Carrero/UConn |
Abstract:
UConn researchers have sequenced the RNA of the most complicated gene known in nature, using a hand-held sequencer no bigger than a cell phone.
A better way to read the genome: If your genome was a library, some genes would be 'Choose Your Own Adventure' novels.
Hartford, CT | Posted on October 12th, 2015If DNA is the blueprint of life, RNA is the construction contractor who interprets it, so sequencing RNA tells you what's really happening inside a cell.
Genomicists Brenton Graveley from the UConn Institute of Systems Genomics, postdoctoral fellow Mohan Bolisetty, and graduate student Gopinath Rajadinakaran teamed up with UK-based Oxford Nanopore Technologies to show that the company's MinION nanopore sequencer can sequence genes faster, better, and at a much lower cost than the standard technology. They published their findings on Sept. 30 in Genome Biology.
If your genome was a library and each gene was a book, some genes would be straightforward reads - but some would be more like a "Choose Your Own Adventure" novel. Researchers often want to know which version of the gene is actually expressed in the body, but for complicated choose-your-own-adventure genes, that has been impossible.
Graveley, Bolisetty, and Rajadinakaran solved the puzzle in two parts. The first was to find a better gene-sequencing technology. In order to sequence a gene using the old, existing technology, researchers first make lots of copies of it, using the same chemistry our bodies use. They then chop up the gene copies into tiny pieces, read each tiny piece, and then, by comparing all the different pieces, try to figure out how they were originally put together. The technique hinges on the likelihood that not all the copies got chopped up into exactly the same pieces. Imagine watching different scenes from a movie, out of order. If you then watched the same movie, but cut into scenes at slightly different places, you could compare the two versions and start to figure out which scenes connect to which.
That technique won't work for choose-your-own-adventure genes, because if you copy them the way the body does, using RNA, each copy can be slightly - or very - different from the next. Such different versions of the same gene are called isoforms. When the different isoforms get chopped up and sequenced, it becomes impossible to accurately compare the pieces and figure out which versions of the gene you started with.
If the gene were a movie, "you wouldn't be able to tell that scenes 1 and 2 were present together," Bolisetty says.
Then last year, the nearly impossible suddenly became possible. Oxford Nanopore, a company based in the UK, released its new nanopore sequencer, and offered one to Graveley's lab. The nanopore sequencer, called a MinION, works by feeding a single strand of DNA through a tiny pore. The pore can only hold five DNA bases - the 'letters' that spell out our genes - at a time. There are four DNA bases, G, A, T, and C, and 1,024 possible five-base combinations. Each combination creates a different electrical current in the nanopore. GGGGA makes a different current than AGGGG, which is different again than CGGGG. By feeding the DNA through the pore and recording the resulting signal, researchers can read the sequence of the DNA.
For the second part of the solution, Graveley, Bolisetty, and Rajadinakaran decided to go big. Instead of sequencing any old choose-your-own-adventure gene, they chose the most complex one known, Down Syndrome cell adhesion molecule 1 (Dscam1), which controls the wiring of the brain in fruit flies. Dscam1 has the potential of making 38,016 possible isoforms, and every fruit fly has the potential to make every one of them, yet how many of these versions are actually made remains unknown.
Dscam1 looks like this: X-12-X-48-X-33-X-2-X, where X's denote sections that are always the same, and the numbers indicate sections that can vary (the number itself shows how many different options there are for that section).
To study how many different isoforms of Dscam1 actually exist in a fly's brain, the researchers first had to convert Dscam1 RNA into DNA. If DNA is the book or set of instructions, RNA is the transcriber that copies the book so that it can be translated into a protein. The DNA includes the instructions for all 38,016 isoforms of the Dscam1 gene, while each individual Dscam1 RNA contains the instructions for just one. No one had yet used a MinION to sequence copies of RNA, and though it was likely it could be done, demonstrating it and showing how well it worked would be a substantial advance in the field.
Rajadinakaran took a fruit fly brain, extracted the RNA, converted it into DNA, isolated the DNA copies of the Dscam1 RNAs, and then ran them through the MinION's nanopores. In this one experiment, they not only found 7,899 of the 38,016 possible isoforms of Dscam1 were expressed but also that many more, if not all versions are likely to be expressed.
"A lot of people said 'The MinION will never work,'" Graveley says, "but we showed it works using the most complicated gene known."
The study demonstrates that gene sequencing technology can now be accessed by a much broader range of researchers than was previously possible, since the MinION is both relatively inexpensive and highly portable so that it requires almost no lab space.
"This type of cutting-edge work puts UConn at the forefront of technology development and strengthens our portfolio of genomics research," says Marc Lalande, director of UConn's Institute for Systems Genomics. "Also, thanks to the investments in genomics through the University's Academic Plan, Brent Graveley can leverage his expertise so that faculty and students across our campuses will successfully compete for grant dollars and launch bioscience ventures."
Graveley will speak about the research at the Oxford Nanopore MinION Community Meeting at the New York Genome Center on Dec. 3.
As for next steps, the researchers plan on going even bigger: sequencing every bit of RNA from beginning to end inside a single cell, something that cannot be done with traditional gene sequencers.
"This technology has amazing potential to transform how we study RNA biology and the type of information we can obtain," says Graveley. "Plus the fact that the MinION is a hand-held sequencer that you plug into a laptop is simply unbelievably cool!"
####
For more information, please click here
Contacts:
Kim Krieger
202-236-0030
Copyright © University of Connecticut
If you have a comment, please Contact us.Issuers of news releases, not 7th Wave, Inc. or Nanotechnology Now, are solely responsible for the accuracy of the content.
| Related News Press |
News and information
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Nanomedicine
![]() New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
![]() New imaging approach transforms study of bacterial biofilms August 8th, 2025
New imaging approach transforms study of bacterial biofilms August 8th, 2025
![]() Cambridge chemists discover simple way to build bigger molecules – one carbon at a time June 6th, 2025
Cambridge chemists discover simple way to build bigger molecules – one carbon at a time June 6th, 2025
![]() Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Discoveries
![]() From sensors to smart systems: the rise of AI-driven photonic noses January 30th, 2026
From sensors to smart systems: the rise of AI-driven photonic noses January 30th, 2026
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Announcements
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Interviews/Book Reviews/Essays/Reports/Podcasts/Journals/White papers/Posters
![]() Metasurfaces smooth light to boost magnetic sensing precision January 30th, 2026
Metasurfaces smooth light to boost magnetic sensing precision January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Tools
![]() Metasurfaces smooth light to boost magnetic sensing precision January 30th, 2026
Metasurfaces smooth light to boost magnetic sensing precision January 30th, 2026
![]() From sensors to smart systems: the rise of AI-driven photonic noses January 30th, 2026
From sensors to smart systems: the rise of AI-driven photonic noses January 30th, 2026
![]() Japan launches fully domestically produced quantum computer: Expo visitors to experience quantum computing firsthand August 8th, 2025
Japan launches fully domestically produced quantum computer: Expo visitors to experience quantum computing firsthand August 8th, 2025
Nanobiotechnology
![]() New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
![]() New imaging approach transforms study of bacterial biofilms August 8th, 2025
New imaging approach transforms study of bacterial biofilms August 8th, 2025
![]() Ben-Gurion University of the Negev researchers several steps closer to harnessing patient's own T-cells to fight off cancer June 6th, 2025
Ben-Gurion University of the Negev researchers several steps closer to harnessing patient's own T-cells to fight off cancer June 6th, 2025
![]() Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
|
|
||
|
|
||
| The latest news from around the world, FREE | ||
|
|
||
|
|
||
| Premium Products | ||
|
|
||
|
Only the news you want to read!
Learn More |
||
|
|
||
|
Full-service, expert consulting
Learn More |
||
|
|
||