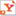Home > Press > Origami: Not just for paper anymore--DNA, folded into complex shapes, could have major impact on nanotechnology
 |
| Mark Bathe, the Samuel A. Goldblith Assistant Professor of Applied Biology Photo: Dominick Reuter |
Abstract:
While the primary job of DNA in cells is to carry genetic information from one generation to the next, some scientists also see the highly stable and programmable molecule as an ideal building material for nanoscale structures that could be used to deliver drugs, act as biosensors, perform artificial photosynthesis and more.
Origami: Not just for paper anymore--DNA, folded into complex shapes, could have major impact on nanotechnology
Cambridge, MA | Posted on April 27th, 2011Trying to build DNA structures on a large scale was once considered unthinkable. But about five years ago, Caltech computational bioengineer Paul Rothemund laid out a new design strategy called DNA origami: the construction of two-dimensional shapes from a DNA strand folded over on itself and secured by short "staple" strands. Several years later, William Shih's lab at Harvard Medical School translated this concept to three dimensions, allowing design of complex curved and bent structures that opened new avenues for synthetic biological design at the nanoscale.
A major hurdle to these increasingly complex designs has been automation of the design process. Now a team at MIT, led by biological engineer Mark Bathe, has developed software that makes it easier to predict the three-dimensional shape that will result from a given DNA template. While the software doesn't fully automate the design process, it makes it considerably easier for designers to create complex 3-D structures, controlling their flexibility and potentially their folding stability.
"We ultimately seek a design tool where you can start with a picture of the complex three-dimensional shape of interest, and the algorithm searches for optimal sequence combinations," says Bathe, the Samuel A. Goldblith Assistant Professor of Applied Biology. "In order to make this technology for nanoassembly available to the broader community — including biologists, chemists, and materials scientists without expertise in the DNA origami technique — the computational tool needs to be fully automated, with a minimum of human input or intervention."
Bathe and his colleagues described their new software in the Feb. 25 issue of Nature Methods. In that paper, they also provide a primer on creating DNA origami with collaborator Hendrik Dietz at the Technische Universitaet Muenchen. "One bottleneck for making the technology more broadly useful is that only a small group of specialized researchers are trained in scaffolded DNA origami design," Bathe says.
DNA consists of a string of four nucleotide bases known as A, T, G and C, which make the molecule easy to program. According to nature's rules, A binds only with T, and G only with C. "With DNA, at the small scale, you can program these sequences to self-assemble and fold into a very specific final structure, with separate strands brought together to make larger-scale objects," Bathe says.
Rothemund's origami design strategy is based on the idea of getting a long strand of DNA to fold in two dimensions, as if laid on a flat surface. In his first paper outlining the method, he used a viral genome consisting of approximately 8,000 nucleotides to create 2-D stars, triangles and smiley faces.
That single strand of DNA serves as a "scaffold" for the rest of the structure. Hundreds of shorter strands, each about 20 to 40 bases in length, combine with the scaffold to hold it in its final, folded shape.
"DNA is in many ways better suited to self-assembly than proteins, whose physical properties are both difficult to control and sensitive to their environment," Bathe says.
Bathe's new software program interfaces with a software program from Shih's lab called caDNAno, which allows users to manually create scaffolded DNA origami from a two-dimensional layout. The new program, dubbed CanDo, takes caDNAno's 2-D blueprint and predicts the ultimate 3-D shape of the design. This resulting shape is often unintuitive, Bathe says, because DNA is a flexible object that twists, bends and stretches as it folds to form a complex 3-D shape.
Written by Anne Trafton, MIT News Office
####
For more information, please click here
Contacts:
Caroline McCall
MIT News Office
E:
T: 617-253-1682
Copyright © MIT
If you have a comment, please Contact us.Issuers of news releases, not 7th Wave, Inc. or Nanotechnology Now, are solely responsible for the accuracy of the content.
| Related News Press |
News and information
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Nanomedicine
![]() New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
![]() New imaging approach transforms study of bacterial biofilms August 8th, 2025
New imaging approach transforms study of bacterial biofilms August 8th, 2025
![]() Cambridge chemists discover simple way to build bigger molecules – one carbon at a time June 6th, 2025
Cambridge chemists discover simple way to build bigger molecules – one carbon at a time June 6th, 2025
![]() Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Sensors
![]() Tiny nanosheets, big leap: A new sensor detects ethanol at ultra-low levels January 30th, 2026
Tiny nanosheets, big leap: A new sensor detects ethanol at ultra-low levels January 30th, 2026
![]() From sensors to smart systems: the rise of AI-driven photonic noses January 30th, 2026
From sensors to smart systems: the rise of AI-driven photonic noses January 30th, 2026
![]() Sensors innovations for smart lithium-based batteries: advancements, opportunities, and potential challenges August 8th, 2025
Sensors innovations for smart lithium-based batteries: advancements, opportunities, and potential challenges August 8th, 2025
Discoveries
![]() From sensors to smart systems: the rise of AI-driven photonic noses January 30th, 2026
From sensors to smart systems: the rise of AI-driven photonic noses January 30th, 2026
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Announcements
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Energy
![]() Sensors innovations for smart lithium-based batteries: advancements, opportunities, and potential challenges August 8th, 2025
Sensors innovations for smart lithium-based batteries: advancements, opportunities, and potential challenges August 8th, 2025
![]() Simple algorithm paired with standard imaging tool could predict failure in lithium metal batteries August 8th, 2025
Simple algorithm paired with standard imaging tool could predict failure in lithium metal batteries August 8th, 2025
Nanobiotechnology
![]() New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
![]() New imaging approach transforms study of bacterial biofilms August 8th, 2025
New imaging approach transforms study of bacterial biofilms August 8th, 2025
![]() Ben-Gurion University of the Negev researchers several steps closer to harnessing patient's own T-cells to fight off cancer June 6th, 2025
Ben-Gurion University of the Negev researchers several steps closer to harnessing patient's own T-cells to fight off cancer June 6th, 2025
![]() Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Solar/Photovoltaic
![]() Spinel-type sulfide semiconductors to operate the next-generation LEDs and solar cells For solar-cell absorbers and green-LED source October 3rd, 2025
Spinel-type sulfide semiconductors to operate the next-generation LEDs and solar cells For solar-cell absorbers and green-LED source October 3rd, 2025
![]() KAIST researchers introduce new and improved, next-generation perovskite solar cell November 8th, 2024
KAIST researchers introduce new and improved, next-generation perovskite solar cell November 8th, 2024
![]() Groundbreaking precision in single-molecule optoelectronics August 16th, 2024
Groundbreaking precision in single-molecule optoelectronics August 16th, 2024
![]() Development of zinc oxide nanopagoda array photoelectrode: photoelectrochemical water-splitting hydrogen production January 12th, 2024
Development of zinc oxide nanopagoda array photoelectrode: photoelectrochemical water-splitting hydrogen production January 12th, 2024
|
|
||
|
|
||
| The latest news from around the world, FREE | ||
|
|
||
|
|
||
| Premium Products | ||
|
|
||
|
Only the news you want to read!
Learn More |
||
|
|
||
|
Full-service, expert consulting
Learn More |
||
|
|
||