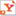Home > Press > INL research probes microbes' potential to clean up groundwater
 |
| Experiments in small glass vials, or microcosms, help reveal the rates at which native bacteria can degrade groundwater pollutants. |
Abstract:
Molecular probes can spotlight cells of specific bacteria species and contaminant-degrading enzymes present in groundwater samples.
By Mike Wall, INL Communications and Governmental Affairs
INL research probes microbes' potential to clean up groundwater
Idaho Falls, ID | Posted on April 16th, 2010In most people's minds, microbes and drinking water don't go well together. Think cholera, or typhoid serious diseases caused by waterborne bacteria. But research at Idaho National Laboratory is showcasing the potential of microbes to cleanse our water rather than foul it.
For the past decade, INL environmental microbiologist Hope Lee has been assessing and tapping this potential. She's pioneering the use of molecular techniques that both identify bacteria present in polluted groundwater and reveal if they're actively breaking the pollutants down. She's finding that naturally occurring microbes can be key cleanup allies, helping degrade contaminants cheaply and effectively.
"I'm just trying to take advantage of what's already down there," Lee says. "I want to make the bacteria work for us."
Clean groundwater: a precious resource
Forty percent of Americans depend entirely or primarily on groundwater to meet basic needs such as drinking and washing. While most U.S. aquifers are clean and safe, chemical contamination poses a threat in many areas. Lee says scientists have documented about 3,000 large underground contaminant "plumes" nationwide, including one at INL's own Test Area North (TAN). These pools of chemicals which can include pesticides, gasoline and diesel fuel, heavy metals and industrial solvents go with groundwater's flow. They can, and sometimes do, end up in people's wells and pipes.
Cleaning up these chemicals is neither quick nor easy. State-of-the-art remediation often centers on a pump-and-treat approach: removing water from the ground and running it through filtration systems. Pump-and-treat works well in many cases, but it's time-consuming and very expensive, sometimes costing tens of thousands of dollars per day.
Lee is looking at ways to supplement remediation technology and potential. Her research could help clean up contaminant plumes more cheaply. The strategy: when possible, shift the heavy lifting from human hands to plume-dwelling microbes.
Gauging bacterial potential
Many naturally occurring bacteria species can degrade contaminants. Sometimes they do it directly, harvesting energy straight from the nasty chemicals. But degradation often occurs accidentally, when the enzymes that microbes use to break down food molecules like methane just happen to degrade contaminants, too.
This latter process is called co-metabolism, and Lee sees much promise in it. She and her colleagues are employing molecular tools to assess the co-metabolic potential of bacteria in contaminant plumes. One of the first steps in this process is a DNA probe of the plume environment.
"We can take a water sample, extract DNA, and tell you what organisms are in there," Lee says.
DNA probes alert researchers to the presence of microbe species known to chew up contaminants. But they can also be tweaked to flag the genes that produce co-metabolizing enzymes. So even if a plume contains some unknown or unfamiliar bacteria species, Lee can at least get an idea of their capabilities.
DNA probes are all about potential; they highlight useful genes, but they're silent on whether or not these genes are being expressed. So Lee uses enzyme activity probes, too. These molecular markers latch onto the degrading enzymes themselves, revealing if plume microbes are actually walking the walk.
Lee integrates probe information with data about the plume's geochemistry also gleaned from water samples to formulate a plan of attack. Say, for example, the probes tell her that several species of aerobic (oxygen-using) bacteria are actively degrading a contaminant. If other measurements indicate the plume is a bit low in oxygen or microbe food, Lee might advise pumping air or methane into the plume to spur bacterial growth.
From potential to practice: How it works
Lee and her colleagues, such as fellow INL microbiologist Mark Delwiche, are some of the first researchers to put these integrated techniques to the test in contaminant plumes around the country. One of their first study sites was TAN, in the sagebrush desert about 50 miles northwest of Idaho Falls.
Parts of TAN contain high levels of trichloroethene, or TCE, perhaps the most common groundwater pollutant in the country. TCE, a phased-out industrial solvent once used to degrease metal equipment, is found at 60 percent of the nation's Superfund sites and in up to 34 percent of U.S. drinking-water supplies. Decades ago, liquid waste containing TCE was injected into a TAN disposal well a common practice at the time, when "the solution to pollution is dilution" was the mantra.
But simple dilution doesn't work as well as people had hoped, so TAN is being remediated. Lee and her team joined the effort. From 2001 to 2003, they took groundwater samples from multiple wells at TAN. They found that TCE concentrations in various parts of the plume were decreasing, suggesting natural microbial attenuation might be occurring. Geochemical measurements revealed that much of the plume contained ample oxygen. And DNA and enzyme activity probes showed that aerobic bacteria were co-metabolically chewing up gobs of contaminant in some portions of the plume.
The probes also gave Lee an idea of the microbial community's composition and health over time. This information is vital, Lee says; her extensive background in community ecology has taught her that microbial cleanup is a team game. Many remediation engineers don't fully appreciate this. They often try to maximize the short-term growth of one or two useful species by injecting air or methane repeatedly.
But multiple species live together and depend on one another, according to Lee, so it could cause problems in the long run if just one or two microbes thrive at the expense of all others.
"You can't feed just one member of a family," she says. "My focus is to keep the whole community happy."
Lee also performed microcosm experiments basically, keeping tabs on vials of contaminated TAN groundwater over time. These studies confirmed that bacteria were breaking TCE down and gave her an idea of the rate at which degradation was proceeding in the field.
With all this evidence in hand, Lee and her colleagues reasoned that aerobic bacteria at TAN were cleaning up some portions of the plume pretty well by themselves. They didn't seem to need big injections of oxygen, methane or any other chemicals. So she said the current strategy of monitored natural attenuation makes sense: let the microbes do most of the work in those parts of the plume and keep an eye on their progress.
This strategy is a big money-saver. Shifting some of the cleanup burden to co-metabolizing microbes while still employing some pump-and-treat and taking advantage of another type of bacterial remediation could substantially cut total costs. And, more importantly, measurements taken last year show that TAN's TCE plume remains under control. The integrated cleanup strategy is working.
Cleaning up groundwater nationwide
Lee has since extended her work, investigating microbial cleanup potential at a half-dozen other sites around the country. By and large, microbes show great promise at all these locations, degrading industrial solvents at relatively rapid rates. Aerobic co-metabolism is often the most efficient and cost-effective route, but in some places, anaerobic bacteria might make the best remediation partners.
"We just try to find the best option for each particular site," Lee says.
Lee is expanding the scope of her research in other ways, too. For example, she's currently studying microbial cleanup processes in soil and clay, not just groundwater. Few people have looked at this problem, focusing instead on groundwater samples because they're easier to work with.
But soil samples could be extremely important, especially when considering the problem of "vapor intrusion." When a contaminant like TCE becomes gaseous, it can diffuse widely, traveling through dirt, clay and even cement building foundations. Lee is now undertaking a study of TCE vapor intrusion at northern Utah's Hill Air Force Base.
The work promises to be difficult, as few previous studies can help guide Lee. But she's not deterred.
"I love a challenge," she says. "And it's so exciting to know that I'm making a difference. I'm helping people clean up the environment."
####
About Idaho National Laboratory
In operation since 1949, INL is a science-based, applied engineering national laboratory dedicated to supporting the U.S. Department of Energy's missions in nuclear and energy research, science, and national defense.
For more information, please click here
Contacts:
Idaho National Laboratory
2525 Fremont Avenue
Idaho Falls, ID 83415
866-495-7440
Copyright © Idaho National Laboratory
If you have a comment, please Contact us.Issuers of news releases, not 7th Wave, Inc. or Nanotechnology Now, are solely responsible for the accuracy of the content.
| Related News Press |
News and information
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Possible Futures
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Announcements
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Environment
![]() Researchers unveil a groundbreaking clay-based solution to capture carbon dioxide and combat climate change June 6th, 2025
Researchers unveil a groundbreaking clay-based solution to capture carbon dioxide and combat climate change June 6th, 2025
![]() Onion-like nanoparticles found in aircraft exhaust May 14th, 2025
Onion-like nanoparticles found in aircraft exhaust May 14th, 2025
Water
![]() Taking salt out of the water equation October 7th, 2022
Taking salt out of the water equation October 7th, 2022
|
|
||
|
|
||
| The latest news from around the world, FREE | ||
|
|
||
|
|
||
| Premium Products | ||
|
|
||
|
Only the news you want to read!
Learn More |
||
|
|
||
|
Full-service, expert consulting
Learn More |
||
|
|
||