Home > Press > Scientists determine the structure of highly efficient light-harvesting molecules in green bacteria
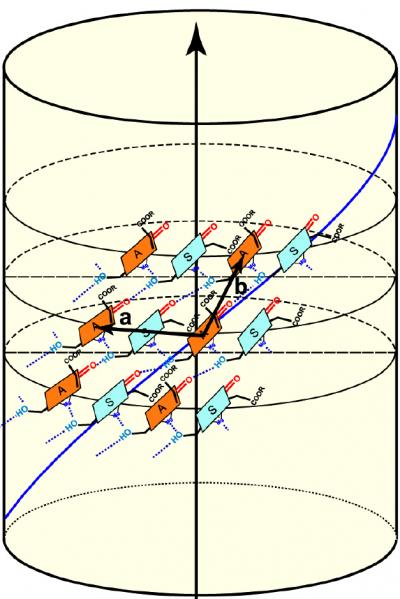 |
| The Nuclear Magnetic Resonance (NMR) results enabled the scientists to determine that the chlorophyll molecules (shown in green and orange) in green bacteria are arranged in helical spirals, and are positioned at an angle to the long axis of the nanotubes.
Credit: Image by Donald Bryant, Penn State University, courtesy of Proceedings of the National Academy of Sciences |
Abstract:
An international team of scientists has determined the structure of the chlorophyll molecules in green bacteria that are responsible for harvesting light energy. The team's results one day could be used to build artificial photosynthetic systems, such as those that convert solar energy to electrical energy. A research paper about the discovery will be published on 4 May 2009 in the Proceedings of the National Academy of Sciences.
Scientists determine the structure of highly efficient light-harvesting molecules in green bacteria
PA | Posted on May 4th, 2009The scientists found that the chlorophylls are highly efficient at harvesting light energy. "We found that the orientation of the chlorophyll molecules make green bacteria extremely efficient at harvesting light," said Donald Bryant, Ernest C. Pollard Professor of Biotechnology at Penn State and one of the team's leaders. According to Bryant, green bacteria are a group of organisms that generally live in extremely low-light environments, such as in light-deprived regions of hot springs and at depths of 100 meters in the Black Sea. The bacteria contain structures called chlorosomes, which contain up to 250,000 chlorophylls. "The ability to capture light energy and rapidly deliver it to where it needs to go is essential to these bacteria, some of which see only a few photons of light per chlorophyll per day."
Because they have been so difficult to study, the chlorosomes in green bacteria are the last class of light-harvesting complexes to be characterized structurally by scientists. Scientists typically characterize molecular structures using X-ray crystallography, a technique that determines the arrangement of atoms in a molecule and ultimately gives information that can be used to create a picture of the molecule; however, X-ray crystallography could not be used to characterize the chlorosomes in green bacteria because the technique only works for molecules that are uniform in size, shape, and structure. "Each chlorosome in a green bacterium has a unique organization," said Bryant. "They are like little andouille sausages. When you take cross-sections of andouille sausages, you see different patterns of meat and fat; no two sausages are alike in size or content, although there is some structure inside, nevertheless. Chlorosomes in green bacteria are like andouille sausages, and the variability in their compositions had prevented scientists from using X-ray crystallography to characterize the internal structure."
To get around this problem, the team used a combination of techniques to study the chlorosome. They used genetic techniques to create a mutant bacterium with a more regular internal structure, cryo-electron microscopy to identify the larger distance constraints for the chlorosome, solid-state nuclear magnetic resonance (NMR) spectroscopy to determine the structure of the chlorosome's component chlorophyll molecules, and modeling to bring together all of the pieces and create a final picture of the chlorosome.
First, the team created a mutant bacterium in order to determine why the chlorophyll molecules in green bacteria became increasingly complex over evolutionary time. To create the mutant, they inactivated three genes that green bacteria acquired late in their evolution. The team suspected that the genes were responsible for improving the bacteria's light-harvesting capabilities. "Essentially, we went backward in evolutionary time to an intermediate state in order to understand, in part, why green bacteria acquired these genes," Bryant said. The team found that the more evolved, wild-type bacteria grow faster at all light intensities than the mutant form. "Indeed, the reason that chlorophylls became more complex was to increase light-harvesting efficiency," said Bryant.
Next, the team isolated chlorosomes from the mutant and the wild-type forms of the bacteria and used cryo-electron microscopy -- a type of electron microscopy that is performed at super-cold cryogenic temperatures -- to take pictures of the chlorosomes. The pictures revealed that chlorophyll molecules inside chlorosomes have a nanotube shape. "They are like Russian dolls, with one concentric tube fitting inside the next," said Bryant. "The mutant bacterium's chlorosomes contain only one set of tubes, whereas the wild-type chlorosomes contain many tubes, each arranged in a unique pattern, like those andouille sausages."
The team then went a step further and used solid-state NMR spectroscopy -- a technique in which samples are spun very rapidly and exposed to a magnetic field -- to look deep inside the chlorosome. This technique enables researchers to understand the relationships between atomic nuclei in a sample and, ultimately, to acquire structural information about the molecules of interest.
"The NMR data revealed to us that the individual chlorophyll molecules in green bacteria are arranged in dimers -- molecules consisting of two identical simpler molecules -- with their long hydrophobic, or water-repellent, tails sticking out of either side," said Bryant. "We also learned precisely how the chlorophyll molecules attach to one another, and we were able to measure the distance between chlorophyll molecules. The cryo-electron microscopy pictures showed gross structural details and distances, and the NMR results allowed us to quantify these distances as well, and confirmed to us that what were were seeing was, in fact, stacks of the chlorophyll molecules all lined up," he said. The NMR results also enabled the scientists to determine that the chlorophyll molecules in green bacteria are arranged in helical spirals. In the mutant bacteria, the chlorophyll molecules are positioned at a nearly 90-degree angle in relation to the long axis of the nanotubes, whereas the angle is less steep in the wild-type organism. "It's the orientation of the chlorophyll molecules that is the most important thing here," said Bryant. The last steps for the team were to pull together all of their data and to create a detailed computer model of the structure.
"At first it seems counterintuitive that green bacteria have managed to evolve a better light-harvesting system by increasing disorder in the chlorosome structure," said Bryant. "Most people would think that if you make something that is more highly ordered, you'll end up with something that works better. But this is clearly a case where that isn't true. If all of the chlorophylls are identically arranged in a chlorosome, then the energy from the photon, once it is absorbed, is going to wander around over all of those chlorophylls, which could take a long time. In the wild-type form, you have these different domains where chlorophyll molecules are located and, therefore, the ability of photon energy to migrate becomes restricted. In other words, the energy in an individual photon visits a smaller number of chlorophylls, and that's an advantage to the organism because the energy can get to where it needs to go faster. Speed is the name of the game that green bacteria play with light. The organisms have only a couple of nanoseconds for the energy to get someplace useful or else the energy is going to be lost. The speed required can be a problem for bacteria that receive only a few photons of light per chlorophyll per day."
Bryant said that the team's results may one day be used to build artificial photosynthetic systems that convert solar energy to electricity. "The interactions that lead to the assembly of the chlorophylls in chlorosomes are rather simple, so they are good models for artificial systems," he said. "You can make structures out of these chlorophylls in solution just by having the right solution conditions. In fact, people have done this for many years; however, they haven't really understood the biological rules for building larger structures. I won't say that we completely understand the rules yet, but at least we know what two of the structures are now and how they relate to the biological system as a whole, which is a huge advance."
The team also includes researchers from the Leiden Institute of Chemistry and the Groningen Biomolecular Sciences and Biotechnology Institute in the Netherlands, and the Max Planck Institute in Germany. This research was supported by the United States Department of Energy.
####
For more information, please click here
Contacts:
Barbara K. Kennedy
814-863-4682
Donald Bryant
814-865-1992
Copyright © Penn State
If you have a comment, please Contact us.Issuers of news releases, not 7th Wave, Inc. or Nanotechnology Now, are solely responsible for the accuracy of the content.
| Related News Press |
News and information
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Discoveries
![]() From sensors to smart systems: the rise of AI-driven photonic noses January 30th, 2026
From sensors to smart systems: the rise of AI-driven photonic noses January 30th, 2026
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Announcements
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Energy
![]() Sensors innovations for smart lithium-based batteries: advancements, opportunities, and potential challenges August 8th, 2025
Sensors innovations for smart lithium-based batteries: advancements, opportunities, and potential challenges August 8th, 2025
![]() Simple algorithm paired with standard imaging tool could predict failure in lithium metal batteries August 8th, 2025
Simple algorithm paired with standard imaging tool could predict failure in lithium metal batteries August 8th, 2025
Solar/Photovoltaic
![]() Spinel-type sulfide semiconductors to operate the next-generation LEDs and solar cells For solar-cell absorbers and green-LED source October 3rd, 2025
Spinel-type sulfide semiconductors to operate the next-generation LEDs and solar cells For solar-cell absorbers and green-LED source October 3rd, 2025
![]() KAIST researchers introduce new and improved, next-generation perovskite solar cell November 8th, 2024
KAIST researchers introduce new and improved, next-generation perovskite solar cell November 8th, 2024
![]() Groundbreaking precision in single-molecule optoelectronics August 16th, 2024
Groundbreaking precision in single-molecule optoelectronics August 16th, 2024
![]() Development of zinc oxide nanopagoda array photoelectrode: photoelectrochemical water-splitting hydrogen production January 12th, 2024
Development of zinc oxide nanopagoda array photoelectrode: photoelectrochemical water-splitting hydrogen production January 12th, 2024
|
|
||
|
|
||
| The latest news from around the world, FREE | ||
|
|
||
|
|
||
| Premium Products | ||
|
|
||
|
Only the news you want to read!
Learn More |
||
|
|
||
|
Full-service, expert consulting
Learn More |
||
|
|
||