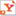Home > Press > Mini DNA sequencer tests true: The performance of the MinION™ miniature DNA sequencing device has been evaluated by an open, international consortium, and the resulting recommendations and protocols published before peer-review on the F1000Research platform
 |
| Oxford Nanopore's MinION™ USB-attached miniature sensing device. CREDIT: Oxford Nanopore Technologies |
Abstract:
Summary:
Public access to Oxford Nanopore's MinION™ miniature sensing device enabled an international consortium to evaluate the technology and provide a standard protocol for its use;
Preliminary analysis of data generated in five very different laboratories indicates the performance and accuracy of the device is consistently good;
Data are freely available for re-analysis and innovation in the Nanopore analysis channel on F1000Research.
Mini DNA sequencer tests true: The performance of the MinION™ miniature DNA sequencing device has been evaluated by an open, international consortium, and the resulting recommendations and protocols published before peer-review on the F1000Research platform
Hinxton, UK | Posted on October 15th, 2015The MinION, a handheld DNA-sequencing device developed by Oxford Nanopore, has been tested and evaluated by an independent, international consortium coordinated by EMBL's European Bioinformatics Institute (EMBL-EBI). The innovative device opens up new possibilities for using sequencing technology in the field, for example in tracking disease outbreaks, testing packaged food or the trafficking of protected species.
The MinION works by detecting individual DNA bases that pass through a nanopore, and unlike existing sequencing technologies, there are few inherent sensing limits on the length of the DNA sequence that it could read at one go. The MinION device was initially made available to thousands of laboratories all over the world, who were inspired to explore the technology and contribute to its development through the MinION Access Programme (MAP).
"The MinION Access Program was a brilliant thing to do. Because the device weighs only 100 grams, Oxford Nanopore could share it easily with more than 1,000 labs worldwide. Some of those people are tinkerers who invented new informatics tools or wet lab techniques that improved MinION performance," says Mark Akeson of the University of California Santa Cruz, a co-inventor of nanopore sequencing, consultant to Oxford Nanopore, and a MAP participant. "The device performs well now, particularly for viral and bacterial genomes, so you can ship it anywhere and know you're going to get the same result. We're looking at a democratisation of sequencing in the not-so-distant future. That is changing things for people who need to solve critical problems in challenging environments, like tracking Ebola strains during the recent outbreak in West Africa. Another challenging environment is space - the MinION will be the first DNA sequencer tested in space, by NASA on the International Space Station."
"In a few years' time, people who may be several steps removed from basic genomic research, like teachers in a classroom, could be using this device to teach science in new, exciting ways that have never been possible before," adds first author Camilla Ip, of Oxford University. "I'm using the MinION in a project with secondary-school students in Oxford because this technology will probably be so much a part of daily life in a few years' time that they take it for granted. The kids who are about to go to university and join the workforce are the ones who will be creating new smartphone apps that use this sensing device for applications we haven't yet imagined."
Five laboratories in the UK, the US, Canada and The Netherlands conducted two sets of ten experiments, for the same E. coli isolate (strain K-12 sub-strain MG1655), using a single, shared protocol. The accuracy and reproducibility of the data were consistent between labs and of good quality. However, there is much work to be done in molecule delivery to flow cells, software protocol clarity and other areas.
The data generated in the study published today represents a snapshot of the MinION's performance in April 2015. Since then, innovation on the MinION has outpaced analysis, with new chips and kits released every 3-6 months. The paper, which includes details of the protocol used and preliminary analysis of the data generated, is available in the Nanopore analysis channel on F1000Research. The data are hosted in the European Nucleotide Archive (ENA).
"Oxford Nanopore has excited the world with the promise of a technology that can be truly disruptive," says David Buck of Oxford University. "When the MinION Access Programme opened it seemed like the largest virtual R&D group ever established in genomics - now the MAP is open to anyone. Working with the MARC team from the start of the MAP has been exciting - we've had the chance to push the technology and gain some traction on understanding what is going on under the hood. We've published the paper in F1000Research before peer review as a baseline for the community, so that everyone can look at the paper and data, go on to do their own analyses share their results and stimulate discussion."
"Nanopore sequencing will open the door for the development of novel tools and applications to analyse the influx of new data," says Rebecca Lawrence, Managing Director of F1000Research. "The Nanopore analysis channel on F1000Research will be a central, open platform on which scientists can publish and discuss new applications and analyse workflows for nanopore sequencing data. People can access and contribute data easily, so that the wider life-science community can realise the full potential of this new technology quickly."
EMBL-EBI has been coordinating the data distribution and analysis for MARC, using the ENA to handle the raw data.
"This new device enables fully mobile sequencing with real-time data streaming, which means that with a high-speed Internet connection, the first dataset could arrive 20 minutes after the DNA is loaded," says Guy Cochrane, who leads the ENA at EMBL-EBI. "The ENA is a public resource that extends the reach and usefulness of sequencing, and with new technologies like this we provide the space, flexibility and expertise needed to test it and get it into shape. We were delighted to use it as the platform for data management and sharing in MAP, so we can place the results in the public domain swiftly."
The next phase of analysis is already underway by the consortium, which is exploring ways to reduce the error rate and pushing to see how small the sample and how long the reads can be.
###
Participating organisations
MAP participants are too numerous to list here. Authors on this paper are from the Wellcome Trust Centre for Human Genetics at the University of Oxford, Queens Medical Centre at the University of Nottingham, The Genome Analysis Centre (TGAC), the University of East Anglia and the European Bioinformatics Institute (EMBL-EBI) in the UK; the Malaghan Institute of Medical Research in New Zealand; the University of British Columbia and the Ontario Institute for Cancer Research in Canada; Virginia Commonwealth University, the University of California Santa Cruz, Brown University and Cold Spring Harbor Laboratory in the US; and ZF-screens B.V. in the Netherlands.
About the MinION Access Programme
The portable MinION device, developed by Oxford Nanopore Technologies Ltd, is a proprietary technology platform for the direct, electronic analysis of single molecules. It works with consumable flow cells that contain the proprietary nanopore sensing apparatus required to perform experiments, and plugs into a PC or laptop that controls the sequencing run. DNA sequencing users generate long reads, analyse data in real-time with a high-speed Internet connection, and use the device in multiple environments. The MinION Access Programme (MAP) is a community-focused technology access programme that started in April 2014 and includes participants in more than 30 countries. Launched as an 'early-access' community the MAP seeks to enable a broad range of people to explore how the MinION may be useful to them in nucleic acid analysis, to contribute to developments in analytical tools and applications and to share their experiences and collaborate. The MAP is now open to any registrant. See tools and publications at publications.nanoporetech.com
About the Nanopore analysis channel
F1000Research, launched in 2013, is an open publishing platform for life scientists that offers immediate publication and transparent peer review. F1000Research allows groups to establish their own channels to serve the needs of their community, for example enabling researchers to publish analyses of nanopore sequencing data quickly and easily. This open-science approach facilitates an effective, global, community-wide discussion around the research being produced. The Nanopore analysis channel published on F1000Research features the Phase 1 MARC paper complete with MinION data and a guest editorial. Guest editors of the Nanopore analysis channel are Nick Loman of the University of Birmingham, Matt Loose of the University of Nottingham, Sara Goodwin of Cold Spring Harbour Library and Hans Jansen of ZF-screens B.V. f1000research.com/channels/nanoporeanalysis
####
About European Bioinformatics Institute
The European Bioinformatics Institute is part of EMBL, and is a global leader in the storage, analysis and dissemination of large biological datasets. EMBL-EBI helps scientists realise the potential of 'big data' by enhancing their ability to exploit complex information to make discoveries that benefit mankind. We are a non-profit, intergovernmental organisation funded by EMBL's 21 member states and two associate member states. Our 570 staff hail from 57 countries, and we welcome a regular stream of visiting scientists throughout the year. We are located on the Wellcome Genome Campus in Hinxton, Cambridge in the United Kingdom.
For more information, please click here
Contacts:
Mary Todd Bergman
44-012-234-94665
Copyright © European Bioinformatics Institute
If you have a comment, please Contact us.Issuers of news releases, not 7th Wave, Inc. or Nanotechnology Now, are solely responsible for the accuracy of the content.
| Related News Press |
Standards/Certifications
![]() NIOSH Releases New Nanotechnology Workplace Design Recommendations March 13th, 2018
NIOSH Releases New Nanotechnology Workplace Design Recommendations March 13th, 2018
![]() Oxford Instruments NanoScience achieves the latest ISO9001:2015 certification March 2nd, 2017
Oxford Instruments NanoScience achieves the latest ISO9001:2015 certification March 2nd, 2017
Govt.-Legislation/Regulation/Funding/Policy
![]() Metasurfaces smooth light to boost magnetic sensing precision January 30th, 2026
Metasurfaces smooth light to boost magnetic sensing precision January 30th, 2026
![]() New imaging approach transforms study of bacterial biofilms August 8th, 2025
New imaging approach transforms study of bacterial biofilms August 8th, 2025
![]() Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Nanomedicine
![]() New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
![]() New imaging approach transforms study of bacterial biofilms August 8th, 2025
New imaging approach transforms study of bacterial biofilms August 8th, 2025
![]() Cambridge chemists discover simple way to build bigger molecules – one carbon at a time June 6th, 2025
Cambridge chemists discover simple way to build bigger molecules – one carbon at a time June 6th, 2025
![]() Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Announcements
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Tools
![]() Metasurfaces smooth light to boost magnetic sensing precision January 30th, 2026
Metasurfaces smooth light to boost magnetic sensing precision January 30th, 2026
![]() From sensors to smart systems: the rise of AI-driven photonic noses January 30th, 2026
From sensors to smart systems: the rise of AI-driven photonic noses January 30th, 2026
![]() Japan launches fully domestically produced quantum computer: Expo visitors to experience quantum computing firsthand August 8th, 2025
Japan launches fully domestically produced quantum computer: Expo visitors to experience quantum computing firsthand August 8th, 2025
Nanobiotechnology
![]() New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
![]() New imaging approach transforms study of bacterial biofilms August 8th, 2025
New imaging approach transforms study of bacterial biofilms August 8th, 2025
![]() Ben-Gurion University of the Negev researchers several steps closer to harnessing patient's own T-cells to fight off cancer June 6th, 2025
Ben-Gurion University of the Negev researchers several steps closer to harnessing patient's own T-cells to fight off cancer June 6th, 2025
![]() Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Alliances/Trade associations/Partnerships/Distributorships
![]() Chicago Quantum Exchange welcomes six new partners highlighting quantum technology solutions, from Chicago and beyond September 23rd, 2022
Chicago Quantum Exchange welcomes six new partners highlighting quantum technology solutions, from Chicago and beyond September 23rd, 2022
![]() University of Illinois Chicago joins Brookhaven Lab's Quantum Center June 10th, 2022
University of Illinois Chicago joins Brookhaven Lab's Quantum Center June 10th, 2022
Research partnerships
![]() Lab to industry: InSe wafer-scale breakthrough for future electronics August 8th, 2025
Lab to industry: InSe wafer-scale breakthrough for future electronics August 8th, 2025
![]() HKU physicists uncover hidden order in the quantum world through deconfined quantum critical points April 25th, 2025
HKU physicists uncover hidden order in the quantum world through deconfined quantum critical points April 25th, 2025
|
|
||
|
|
||
| The latest news from around the world, FREE | ||
|
|
||
|
|
||
| Premium Products | ||
|
|
||
|
Only the news you want to read!
Learn More |
||
|
|
||
|
Full-service, expert consulting
Learn More |
||
|
|
||