Home > Press > Nanobotmodels offer detailed model of nuclear pore complex
 |
Abstract:
Nuclear pores are large protein complexes that cross the nuclear envelope, which is the double membrane surrounding the eukaryotic cell nucleus. There are about an average of 2000 nuclear pore complexes (NPCs), in the nuclear envelope of a vertebrate cell, but it varies depending on cell type and the stage in the life cycle.
Nanobotmodels offer detailed model of nuclear pore complex
Melitopol, Ukraine | Posted on August 11th, 2014Small particles (< ~40 kDa) are able to pass through the nuclear pore complex by passive diffusion. Larger particles are also able to pass through the large diameter of the pore but at almost negligible rates. Efficient passage through the complex requires several protein factors. Karyopherins, which may act as import ins or export ins are part of the Importin-β super-family which all share a similar three-dimensional structure.
There are different export pathways through the NPC for each RNA class that exists. RNA export is also signal mediated (NES); the NES is in RNA-binding proteins (except for tRNA which has no adapter). It is notable that all viral RNAs and cellular RNAs (tRNA, rRNA, U snRNA, microRNA) except mRNA are dependent on RanGTP. Conserved mRNA export factors are necessary for mRNA nuclear export.
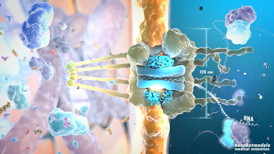
Nanobotmodels Medical Animation (www.nanobotmodels.com) made detailed model of nuclear pore complex. Picture of the nuclear pore cross-section shows viral RNA transport into the nucleus. This model is the part of cancer education project and will cover human papillomavirus life cycle.
Nanobotmodels made this detailed model of nuclear pore and surrounding proteins uses ePMV software to show real protein structure. The open-source uPy plugin, embedded Python Molecular Viewer (ePMV) runs molecular-modeling software directly in several professional 3D animation applications.
####
For more information, please click here
Copyright © Nanobotmodels Company
If you have a comment, please Contact us.Issuers of news releases, not 7th Wave, Inc. or Nanotechnology Now, are solely responsible for the accuracy of the content.
| Related News Press |
News and information
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Nanomedicine
![]() New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
![]() New imaging approach transforms study of bacterial biofilms August 8th, 2025
New imaging approach transforms study of bacterial biofilms August 8th, 2025
![]() Cambridge chemists discover simple way to build bigger molecules – one carbon at a time June 6th, 2025
Cambridge chemists discover simple way to build bigger molecules – one carbon at a time June 6th, 2025
![]() Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Announcements
![]() Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
Decoding hydrogen‑bond network of electrolyte for cryogenic durable aqueous zinc‑ion batteries January 30th, 2026
![]() COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
COF scaffold membrane with gate‑lane nanostructure for efficient Li+/Mg2+ separation January 30th, 2026
Nanobiotechnology
![]() New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
New molecular technology targets tumors and simultaneously silences two ‘undruggable’ cancer genes August 8th, 2025
![]() New imaging approach transforms study of bacterial biofilms August 8th, 2025
New imaging approach transforms study of bacterial biofilms August 8th, 2025
![]() Ben-Gurion University of the Negev researchers several steps closer to harnessing patient's own T-cells to fight off cancer June 6th, 2025
Ben-Gurion University of the Negev researchers several steps closer to harnessing patient's own T-cells to fight off cancer June 6th, 2025
![]() Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
Electrifying results shed light on graphene foam as a potential material for lab grown cartilage June 6th, 2025
|
|
||
|
|
||
| The latest news from around the world, FREE | ||
|
|
||
|
|
||
| Premium Products | ||
|
|
||
|
Only the news you want to read!
Learn More |
||
|
|
||
|
Full-service, expert consulting
Learn More |
||
|
|
||